| Platforms | OS | R CMD check | Coverage |
|---|---|---|---|
| Travis CI | Linux | ||
| Bioc (devel) | Multiple | NA |
|
| Bioc (release) | Multiple | NA |
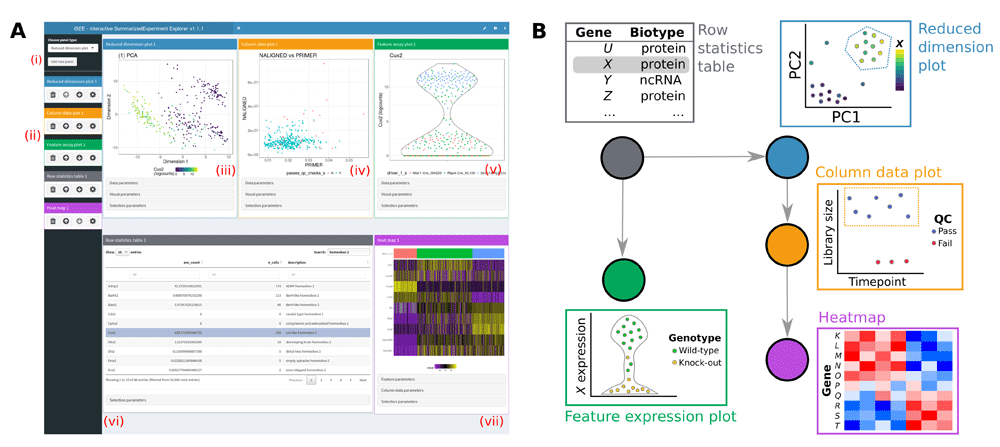
The iSEE package provides an interactive user interface for exploring data in objects derived from the SummarizedExperiment class.
Particular focus is given to single-cell data stored in the SingleCellExperiment derived class.
The user interface is implemented with RStudio's Shiny, with a multi-panel setup for ease of navigation.
This initiative was proposed at the European Bioconductor Meeting in Cambridge, 2017. Current contributors include:
The user interface of iSEE web-applications currently offers the following features:
✅ Multiple interactive plot types with selectable points.
✅ Interactive tables with selectable rows.
✅ Coloring of samples and features by metadata or expression data.
✅ Zooming to a plot subregion.
✅ Transmission of point selections between panels to highlight, color, or restrict data points in the receiving panel(s).
✅ Lasso point selection to define complex shapes.
The iSEE user interface currently contains the following components where each data point represents a single biological sample:
✅ Reduced dimension plot: Scatter plot of reduced dimensionality data.
✅ Column data plot: Adaptive plot of any one or two sample metadata. A scatter, violin, or square design is dynamically applied according to the continuous or discrete nature of the metadata.
✅ Feature assay plot: Adaptive plot of expression data across samples for any two features or one feature against one sample metadata.
✅ Column statistics table: Table of sample metadata.