TransCorpus is a scalable, production-ready API and CLI toolkit for large-scale parallel translation, preprocessing, and corpus management. It supports multi-GPU translation, robust checkpointing, and safe concurrent downloads, making it ideal for research and industry-scale machine translation workflows.
- 🚀 Multi-GPU and multi-process translation
- 📦 Corpus downloading and preprocessing
- 🔒 Safe, resumable, and concurrent file downloads
- 🧩 Split and checkpoint management for large corpora
- 🛠️ Easy deployment and extensibility
- 🖥️ Cross-platform: Linux, macOS, Windows
- Clone and Install
git clone https://github.com/jknafou/TransCorpus.git
cd TransCorpus
UV_INDEX_STRATEGY=unsafe-best-match rye sync
source .venv/bin/activate- Download a Corpus
transcorpus download-corpus [corpus_name]- Preprocess the corpus by splits
transcorpus preprocess [corpus_name] [language] --num-split 100- Translate (and preprocess if not done) the corpus by split
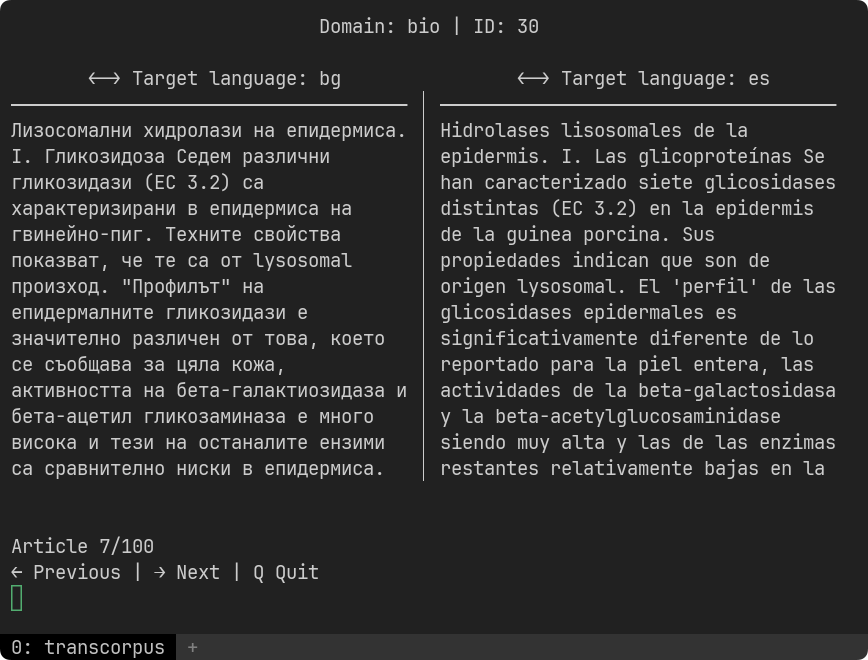
transcorpus translate [corpus_name] [language] --num-split 100- Preview a corpus with two languages next to each other:
transcorpus preview [corpus_name] [language1] Opt[language2]A demo mode can be tested using the -d flag for each command.
The following example translates the bio corpus (PubMed) of about 30GB, preprocessing it with 4 parallel workers, while translating each available split with two GPUs of different sizes. It can easily be modified to one needs. When deployed on an HPC cluster, for example with SLURM, it will automatically resume from where it left off in the previous run. With shared memory, multiple GPUs from different nodes can work simultaneously.
# Preprocess with 4 workers iteratively, split into 20 parts (here in demo mode)
./example/multi_GPU.sh bio de 4 20
TransCorpus enables the training of state-of-the-art language models through synthetic translation. For example, TransBERT achieved superior performance by leveraging corpus translation with this toolkit. A paper detailing these results will be submitted to EMNLP 2025. 📝 Current Paper Version. If you use this toolkit, please cite:
@misc{knafou-transbert,
author = {Knafou, Julien and Mottin, Luc and Ana\"{i}s, Mottaz and Alexandre, Flament and Ruch, Patrick},
title = {TransBERT: A Framework for Synthetic Translation in Domain-Specific Language Modeling},
year = {2025},
note = {Submitted to EMNLP2025. Anonymous ACL submission available:},
url = {https://transbert.s3.text-analytics.ch/TransBERT.pdf},
}
Looking for pretrained models built with TransCorpus? Check out TransBERT-bio-fr on Hugging Face 🤗, a French biomedical language model trained entirely on synthetic translations generated by this toolkit. Also available, TransCorpus-bio-fr on Hugging Face 🤗
One can easily add its own corpus (along with a demo) to the repo following the same schema of domains.json:
"bio": {
"database": {
"file": "https://transcorpus.s3.text-analytics.ch/bibmed.tar.gz"
},
"corpus": {
"file":
"https://transcorpus.s3.text-analytics.ch/title_abstract_en.txt"
,
"demo":
"https://transcorpus.s3.text-analytics.ch/1k_sample.txt"
},
"id": {
"file":
"https://transcorpus.s3.text-analytics.ch/PMID.txt"
,
"demo":
"https://transcorpus.s3.text-analytics.ch/PMID_1k_sample.txt"
},
"language": "en"
}Where each line of the corpus is a different document. For the moment, a life-science corpus is available comprising about 28GB of raw text, 22M of abstracts from PubMed. The database it is made of can also be downloaded using transcorpus download-database bio.
- Python 3.10+
- rye (for dependency management)
- CUDA-enabled GPUs (for multi-GPU translation)
Pull requests and issues are welcome!
MIT License
- Swiss AI Center
- fairseq
- PyTorch
- rye
TransCorpus makes large-scale, robust translation easy and reproducible.